About 3 months ago, I got a 23AndMe kit, spit into a tube and sent my biosample to the 23AndMe lab to have my genome sequenced. I’m obsessed with creating infographics and so I had this vision that I could take my genetic data and look at it in a new way, maybe create art out of it. And so, for the last 3 months, I’ve spent my evenings hacking away, trying to understand the data I got and what I can do with it.
This is my first truly public visualization, but it’s not of my personal genetic data. This is a visualization of the 300 most researched SNPs, sorted by location in the genome, colored by chromosome placement, and scaled by “importance” (the amount of research done on each SNP).
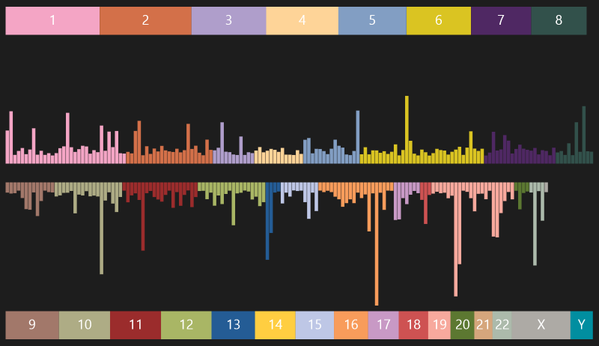
If that last sentence made very little sense, it’s because you’re where I was 3 months ago.
When my raw genetic data came back from 23AndMe, I struggled immensely to try to understand the context of the raw data with which I was playing. I wished there was a simple resource to helping people with a non-genetics background and education understand the basics of what data and resources are available and how to start working with that data as quickly as possible to start trying to understand and explore the incredible amount of data out there.
Hopefully, this is that guide. My goal here will be to give you enough information to start playing with the data. To say that genetics is a large complex field is like saying the sun is hot-ish. Here I just want to introduce some terms so you can feel like you have some idea what you’re looking at when you look at raw genetic data.
Note that I’m not a geneticist. This piece represents what I’ve learned (or perhaps mislearned) through a few months of data munging, confused research, and many questions to people who know more than I do.
I’ll start where I started (my 23AndMe data) and work backwards from there. If you’re not familiar with 23AndMe, it is a genome sequencing and research company. Their consumer-facing product is a $199 genome sequencing kit. Pay the money, spit in the tube, wait for the sequencing, and they’ll send you an ancestral analysis, connect you with others who share your heritage (I found mostly 3rd and 4th cousins), and let you browse or download your “raw data” in the form of your alleles for 1.5 million SNPs (pronounced “snips”).
UPDATE: Thanks to FDA approval, 23AndMe now also provides carrier status for some genetic diseases, a genetics wellness report, and a traits report. So go get it done now!
Not interested in waiting? Lucky you! There is a website called OpenSNP where you can download someone else’s raw data. Now you have millions of SNPs to work with. They’ll have exciting names like “rs6313” and exciting data like “GG” or “AG” or “CT”.
Wait. What? Back up.
Trying to learn about genetics by starting with a pile of SNPs is like trying to learn how to drive by trying to put an engine back together. To even understand what this data signifies, we have to back up. A lot. Let’s start with the basics. We’ll go in this order: DNA -> chromosomes -> genes -> SNPs.
Everyone has at least heard of DNA. A DNA (deoxyribonucleic acid) molecule is essentially a code for building living things. You have DNA, animals have DNA, trees have DNA, bacteria have DNA. This code is written out in a sequence of 4 letters which represent the 4 chemical bases in a DNA molecule: AGCT. If you forget, just remember that Gattaca is spelled using just the letters that make up DNA.
You can literally read this code out in a sequence, which will look something like this:
GTTCAGGCAAAGGCAGCAGTTGCTAATGAAGACACTGGAGGACAGCGGGCTT
That is an actual sequence from the USCS Genome browser, which hosts the full human genome and which is a powerful tool for confusing someone who is just trying to understand the basics.
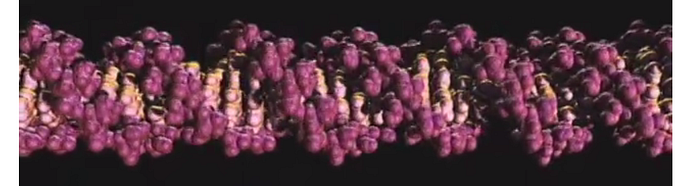
The DNA structure is a double helix. This means that, when it comes time for a cell to split and the DNA to replicate, it “unzips” along the center to form 2 half-strands of DNA.
The half-strand becomes complete again by binding to the appropriate chemicals. Each A in the sequence will bind to a T and each G will bind to a C. This is why, even though the DNA is a double-helix, we only need to know one side. The other side of the sequence can only be the corresponding letter.
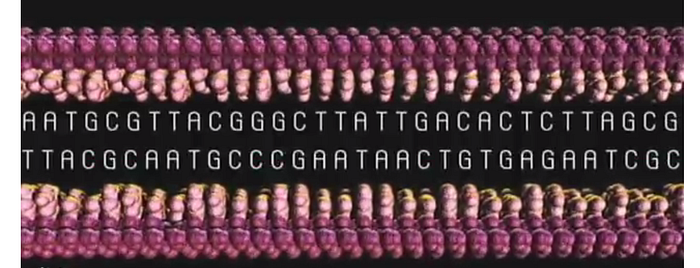
And because these letters must go together, we call each individual letter a “base pair”.
The human genome consists of about 3.2 billion of these base pairs.
Chromosomes
So where is all the information contained? Unless you are terribly unlucky, you have 23 chromosomes. Actually… you have 46 chromosomes, 23 from your mom and 23 from your dad. These chromosomes are autosomes numbered 1 to 22 plus the sex chromosome, which is XX for females and XY for males.
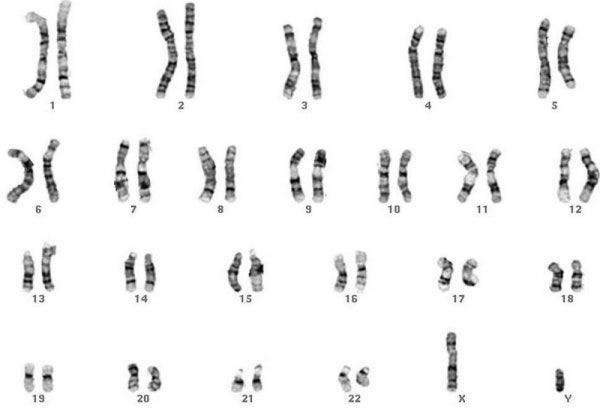
When I say “you” have these chromosomes, I mean your cells have them. Inside the nucleus of the majority of the cells in your body sit these chromosomes and when your cells divide, your chromosomes pack themselves up, split, and head over to another cell.
Each chromosome has tens of millions of base pairs and the lower-numbered ones tend to be longer than the higher numbered ones. If you go back to the visual above, the colored chromosome numbers are scaled according to how many base pairs each chromosome has.
Genes
Within each chromosome are a sequence of genes. Genes are sequences of these base pairs that will result in encoding proteins in our bodies. The result of these protein encoding is… you.
As an example, different populations can carry certain mutations of a gene. One of my favorite examples (because it is so stark) is the BRCA1 mutation. Located (I think) on chromosome 17, this gene correlates with an 80–90% chance of getting breast cancer.
A mother****ing 80–90% chance. It’s estimated that 5–10% of all breast cancer cases are due to this gene mutation that only shows up in 0.3% of all people. This is why carriers of this extremely dangerous mutation (like Angelina Jolie) may opt to undergo a preventative mastectomy. This is a case of personal genomic data saving lives.
When there is a line between a genetic trait and some physical manifestation of that trait, it is called a phenotype. The “classic” example of a phenotype is eye color.
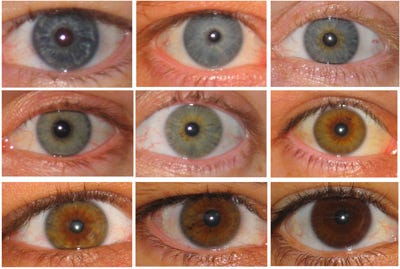
Although eye color is clearly a genetic trait, drawing a direct line between gene mutations and eye color can be very difficult because as many as 16 different genes may play different roles.
More Alike Than Different
“Wait, if there are 3.2 billion base pairs in human DNA, why do I only have 1.5 million data points” you may say. The answer is that for the vast, vast majority of DNA, humans are all exactly the same. Something higher than 99% of our genomes. Like… 99.8%.

Or more. I’m looking for the exact number, but it’s hard to find. The fact of the matter is that it would be pointless to test for data where the results are always going to come out the same way.
Fortunately, geneticists have identified a huge number of spots in the human genome where different people have different information. These locations are called single-nucleotide polymorphisms or…
Wait for it…
SNPs
The 1.5 million SNPs delivered to me by 23AndMe that I have to play with are surgically identified sites in the human genome where there are likely to be differences between different people. Each SNP is identified with a unique name and is identified (usually) as a set of two A, G, C, or T at a specific position within a chromosome.
So why, when you look at your data at a specific single location, do you have two letters? Each letter is from one copy of the chromosomes that you have, one from mom and one from dad. If you’re male, you’ll see that you only have one letter in the SNPs that are in the Y and X chromosome. That’s because you didn’t get two copies of the X chromosome against which we could compare.
But What Does It Mean?!?
SNPs are essential to genetic research. They allow scientists to look at the unique parts of the genome instead of the same-y parts when doing phenome analysis, disease determination, or drug resistance research. They help researchers (relatively) quickly determine the probabilities that certain mutations are associated with certain traits. It is not necessarily true that a given SNP result is *causing* a given trait, but rather that it is statistically associated with it.
Let’s go back to our BRCA1 gene mutation. Identifying this mutation isn’t as simple as looking at a single place in the genome and saying “there it is!” There are scores of SNPs that have been identified as associated with this mutation. BRAC1 is an incredibly stark example of how personal genetics can tell us more about our health. There are many other genes and conditions where the associations aren’t nearly as clear and the consequences far less certain.
SNPs are where you’ll find most of the data around personal genome research. That is why the data you download refers to SNPs and why genetics research talks about SNPs and why things like OpenSNP and SNPedia are such valuable resources for doing data-centered research. And that is why a visualization of the 300 most researched SNPs is so interesting… because it tells us where in the genome researchers are looking for data.
Give Me More!
If you’re interested in more high-level information about genetics, there are two books that got me excited about genetic data in the first place. Interestingly, both of these authors started not as biologists but as physicists, which is where my initial love of science took hold.
The Human Genome by John Quackenbush

This is a great, fast introduction to the human genome. It requires repeat readings because it is over-packed with information and terminology. I loved Quackenbush’s writing style & his structured intro to the field.
Language of God by Francis Collins

This might be a bit of a special interest book. Francis Collins is the director of the NIH and, godfather of the Human Genome Project, and part of the team that first identified the BRCA mutation as it relates to breast cancer. He’s also a committed Christian. As a Christian, I was fascinated to read Collins’ overview of how his faith and his genetics research intertwine. The book is part philosophy, part genetics introduction, part historical record of the race for the Human Genome Project. If you’re not a Christian, I still recommend it just for his fascinating and deeply personal portrayal of some of the most important moments in the history of genetics.
Data Resources
23AndMe API — If you’re interested in building apps against the 23AndMe data, they have an API that you can build for. I haven’t published anything using their API yet, if you sign in (it’s free) you can get a list of all the SNPs that 23AndMe encodes for.
OpenSNP — This is my hub for research. Tons of data free for download, including many actual 23AndMe data sets. This is where I got my data on how much research has been done for any given SNP. The OpenSNP ranking is based on entries in the SNPedia, papers published in PLoS and Genome.gov, annotations in the Personal Genome Project and papers on Mendeley. Plus you can download their full annotation dump at one go so you can see all the details that they base their rankings on. It’s amazing.
SNPedia — This is an open wiki for SNP associations. It has an enormous amount of information on specific SNPs, the different variations and their associations with phenotypes and medical conditions. It doesn’t have the raw data files available through OpenSNP, but it has far more detail on the associations of individual SNPs.
UCSC Genome Browser — I gave the UCSC genome browser a hard time earlier, but that’s because it was the first place I went and, if you’re not already a geneticist, it is going to be hard to get any helpful information out of it. However, it does some amazing things with cross-species comparative genomics and they host raw data downloads for the genomes of dozens of different species.